An open-source R/Bioconductor package for tabular omics data analysis using
a supra-hexagonal map
News
- The artwork called supraHex has won The Best Artwork Award in ISMB 2014. This artwork is automatically done and is reproducible here.
- Demonstrated in a variety of genome-wide datasets such as:
leukemia patient transcriptome, time-course transcriptome, RNA-seq data, DNA replication timing, cell type evolution, and clonal population structure.
Introduction
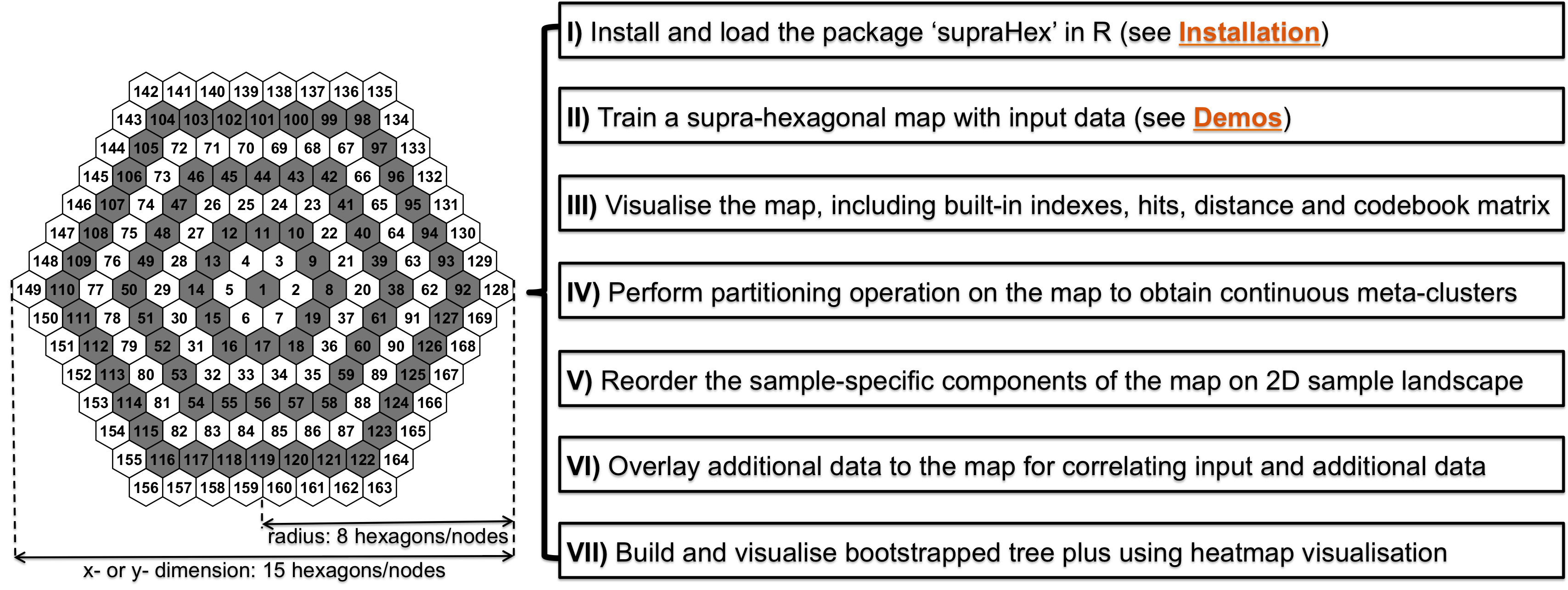
The supra-hexagonal map is a giant hexagon on a 2-dimensional map grid seamlessly consisting of smaller hexagons.
supraHex intends to meet the need for quickly understanding genome-wide biological data, which usually involve a large number of genomic coordinates (e.g. genes) but a much smaller number of samples.
supraHex first uses a supra-hexagonal map to self-organise the input omics data, and then post-analyses the trained map for integrated tasks: simultaneous analysis of genes and samples, and multilayer omics data comparisons.
supraHex aims to deliver an eye-intuitive tool and a dedicated website with extensive online documentation and easy-to-follow demos.
For more, see slides and poster in ISMB2014.
Features
- The supra-hexagonal map trained via a self-organising learning algorithm;
- Visualisations at and across nodes of the map;
- Partitioning of the map into gene meta-clusters;
- Sample correlation on 2D sample landscape;
- Overlaying additional data onto the trained map for exploring relationships between input and additional data;
- Support for heatmap and tree building and visualisations;
- Used by the package dnet for network-based sample classifications;
- This package can run on
Windows,MacandLinux.
Workflow
Manual
Demos
- Demo for multilayer omics dataset from Hiratani et al
- Demo for leukemia patient dataset from Golub et al
- Demo for human embryo dataset from Fang et al
- Demo for arabidopsis embryo dataset from Xiang et al
- Demo for for the artwork in ISMB 2014
- Demo for human cell type evolutionary profile dataset from Sardar et al
- Demo for postprocessing cellular prevalence of mutations in clonal populations (inferred by PyClone)
- Demo for analysing RNA-seq dataset (along with edgeR)
URL
Dependency
- Depends: hexbin
- Imports: ape, MASS, grDevices, graphics, stats, utils
- Suggests:
- Extends: