Start at 2017-03-27 18:59:40
First, define topology of a map grid (2017-03-27 18:59:40)...
Second, initialise the codebook matrix (61 X 10) using 'linear' initialisation, given a topology and input data (2017-03-27 18:59:40)...
Third, get training at the rough stage (2017-03-27 18:59:40)...
1 out of 7 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
2 out of 7 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
3 out of 7 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
4 out of 7 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
5 out of 7 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
6 out of 7 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
7 out of 7 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
Fourth, get training at the finetune stage (2017-03-27 18:59:40)...
1 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
2 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
3 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
4 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
5 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
6 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
7 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
8 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
9 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
10 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
11 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
12 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
13 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
14 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
15 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
16 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
17 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
18 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
19 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
20 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
21 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
22 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
23 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
24 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
25 out of 25 (2017-03-27 18:59:40)
updated (2017-03-27 18:59:40)
Next, identify the best-matching hexagon/rectangle for the input data (2017-03-27 18:59:40)...
Finally, append the response data (hits and mqe) into the sMap object (2017-03-27 18:59:40)...
Below are the summaries of the training results:
dimension of input data: 100x10
xy-dimension of map grid: xdim=9, ydim=9, r=5
grid lattice: hexa
grid shape: suprahex
dimension of grid coord: 61x2
initialisation method: linear
dimension of codebook matrix: 61x10
mean quantization error: 4.60312441206232
Below are the details of trainology:
training algorithm: batch
alpha type: invert
training neighborhood kernel: gaussian
trainlength (x input data length): 7 at rough stage; 25 at finetune stage
radius (at rough stage): from 3 to 1
radius (at finetune stage): from 1 to 1
End at 2017-03-27 18:59:40
Runtime in total is: 0 secs
Start at 2017-03-27 18:59:40
First, define topology of a map grid (2017-03-27 18:59:40)...
Second, initialise the codebook matrix (30 X 61) using 'linear' initialisation, given a topology and input data (2017-03-27 18:59:40)...
Third, get training at the rough stage (2017-03-27 18:59:40)...
1 out of 300 (2017-03-27 18:59:40)
30 out of 300 (2017-03-27 18:59:40)
60 out of 300 (2017-03-27 18:59:40)
90 out of 300 (2017-03-27 18:59:40)
120 out of 300 (2017-03-27 18:59:40)
150 out of 300 (2017-03-27 18:59:40)
180 out of 300 (2017-03-27 18:59:40)
210 out of 300 (2017-03-27 18:59:40)
240 out of 300 (2017-03-27 18:59:40)
270 out of 300 (2017-03-27 18:59:40)
300 out of 300 (2017-03-27 18:59:40)
Fourth, get training at the finetune stage (2017-03-27 18:59:40)...
1 out of 1200 (2017-03-27 18:59:40)
120 out of 1200 (2017-03-27 18:59:40)
240 out of 1200 (2017-03-27 18:59:40)
360 out of 1200 (2017-03-27 18:59:40)
480 out of 1200 (2017-03-27 18:59:40)
600 out of 1200 (2017-03-27 18:59:40)
720 out of 1200 (2017-03-27 18:59:40)
840 out of 1200 (2017-03-27 18:59:40)
960 out of 1200 (2017-03-27 18:59:40)
1080 out of 1200 (2017-03-27 18:59:40)
1200 out of 1200 (2017-03-27 18:59:40)
Next, identify the best-matching hexagon/rectangle for the input data (2017-03-27 18:59:40)...
Finally, append the response data (hits and mqe) into the sMap object (2017-03-27 18:59:40)...
Below are the summaries of the training results:
dimension of input data: 10x61
xy-dimension of map grid: xdim=6, ydim=5, r=3
grid lattice: rect
grid shape: sheet
dimension of grid coord: 30x2
initialisation method: linear
dimension of codebook matrix: 30x61
mean quantization error: 4.0169186787623
Below are the details of trainology:
training algorithm: sequential
alpha type: invert
training neighborhood kernel: gaussian
trainlength (x input data length): 30 at rough stage; 120 at finetune stage
radius (at rough stage): from 1 to 1
radius (at finetune stage): from 1 to 1
End at 2017-03-27 18:59:40
Runtime in total is: 0 secs
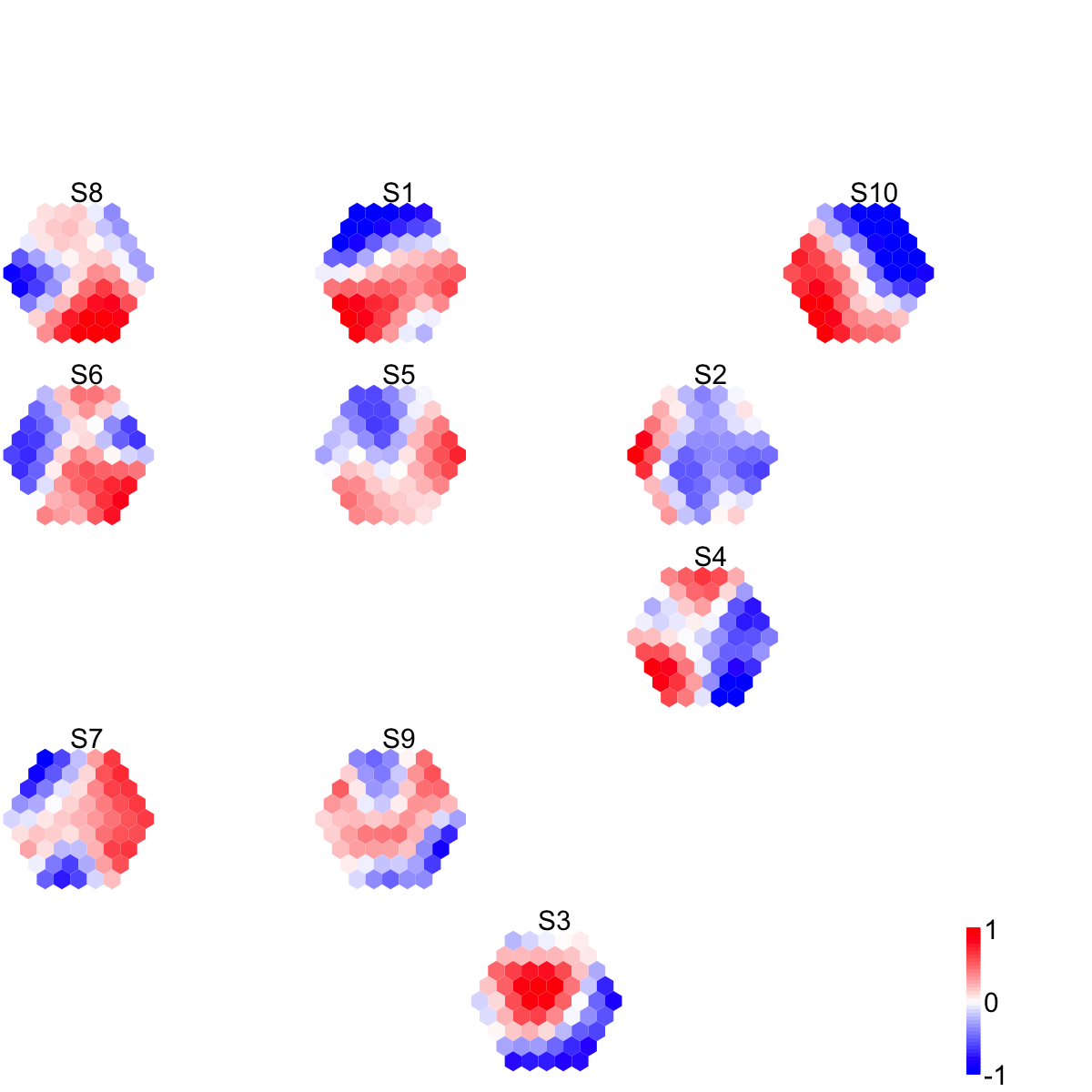
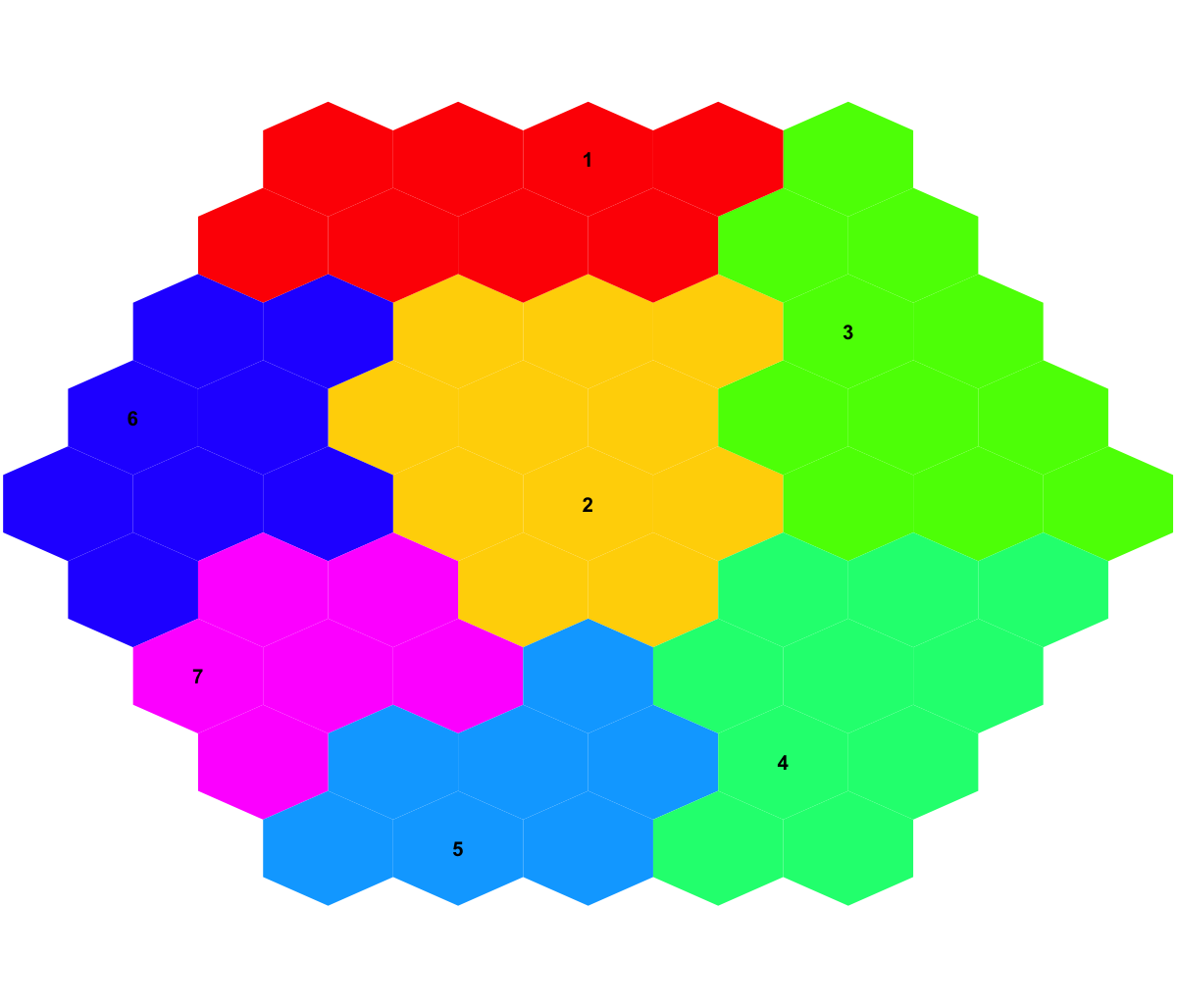
 visDmatCluster(sMap,sBase)
# As you have seen the previous two images, not only can you tell sample relationships but also the meta-clusters wherein.
visDmatCluster(sMap,sBase)
# As you have seen the previous two images, not only can you tell sample relationships but also the meta-clusters wherein.
